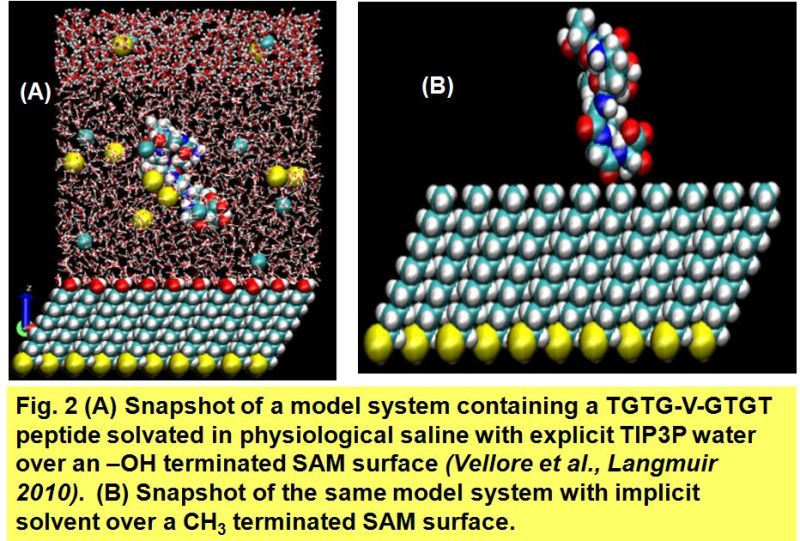
Implicit Solvation Effect of Peptide Interaction with Hydrophobic and Hydrophilic SAMs
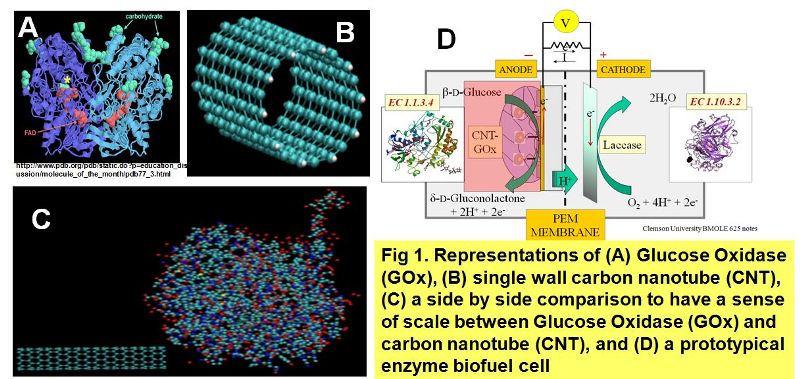
PosterStatistics from the North Central Regional Trauma Advisory Council have shown that trauma is the national leading cause of death for people between the ages of 1 and 44 in the United States. Trauma, which includes hemorrhaging from automobile crashes, accounts for 62% of all deaths for young adults between the ages of 15 and 24. More work years are lost due to trauma than cancer and heart disease combined. In the last 50 years, not much has been done to improve trauma management at the site of injury. As such, there is the need to develop fully implantable biosensors with high stability and performance for use in trauma management at the site of injury. Implantable biofuel cells (Fig. 1d) offer an attractive alternative for powering fully implantable biomedical devices.
A proposed implantable biosensor has been developed by Guiseppi et al. that makes use of the immobilization of a noncovalent conjugate system onto a working electrode. The noncovalent conjugate system consists of the ultrasonic processing of carbon nanotubes (Fig. 1b) and glucose oxidase (Fig. 1a). The orientation and conformation of the enzyme around the carbon nanotubes is essential to obtaining a high performance and stable biosensor. We will like for the redox active site (flavin adenine dinucleotide) to be proximal to the carbon nanotubes to improve the electron transfer mechanism thus increasing the biosensor's sensitivity.
In order to simulate this large conjugate system, there is the need to develop the appropriate modeling parameters. To do this, our initial model system includes a nine amino acid peptide (TGTG-V-GTGT) interacting with a CH3-terminated self-assembled monolayer (SAM) surface and an OH-terminated SAM surface (Fig. 2). Solvation effects will be simulated implicitly using the Generalized Born with a simple Switching (GBSW) method. The CHARMM suite of simulation tools is being used as a simulation engine conducted with the CHARMM force field. Once the appropriate parameters have been developed, the simulation will be extended to the interaction of the TGTGVGTGT peptide with various functionalized carbon nanotubes of different lengths.
_________________________________________________________________
Joint advised with Dr. Anthony Guiseppi-Elie, Dept. Chemical & Biological Engr.