Molecular Dynamics Simulation of Structured Peptide Adsorption to Functionalized Self-Assembled Monolayer Surfaces
PosterImprovements in the performance of computational hardware and simulation techniques have made molecular dynamics (MD) simulations of large, solvated protein systems sufficiently tractable to warrant the use of this approach in studying the mechanistic details of protein adsorption. The goal of this project was to evaluate the applicability of existing parameter libraries (CHARMM, AMBER, and OPLS-AA)for representing the various aspects of protein-surface interactions. We simulated discrete protein structural components in the form of simple model peptide structures (alpha-helices and beta-sheets), as they interact with hydrophobic and charged self-assembled monolayer (SAM) surfaces. These simulations were conducted as a collaborative effort in parallel with experimental studies (ssNMR, XPS, ToF-SIMS, NEXAFS) of similar and identical peptide-surface systems by collaborators at the University of Washington, Seattle, WA. Our simulations employed the replica-exchange molecular dynamics (REMD) technique for optimizing the peptide conformational search. These simulations included explicitly solvated systems approximating 140 mM (physiological) saline. Structural analyses included peptide secondary structure studies, orientation studies, as well as various interatomic distance and side-chain tilt-angle measurements. Solution structure analyses included water dipole orientations, water and ion density distributions, and diffusion characteristics. The structural information resulting from these simulations was compared to results from the experimental studies to both validate the theoretical approach used and to serve as a basis for comparing the performance of various force fields. Results showed that the CHARMM force field provided results that were most consistent with experimentally measured values.
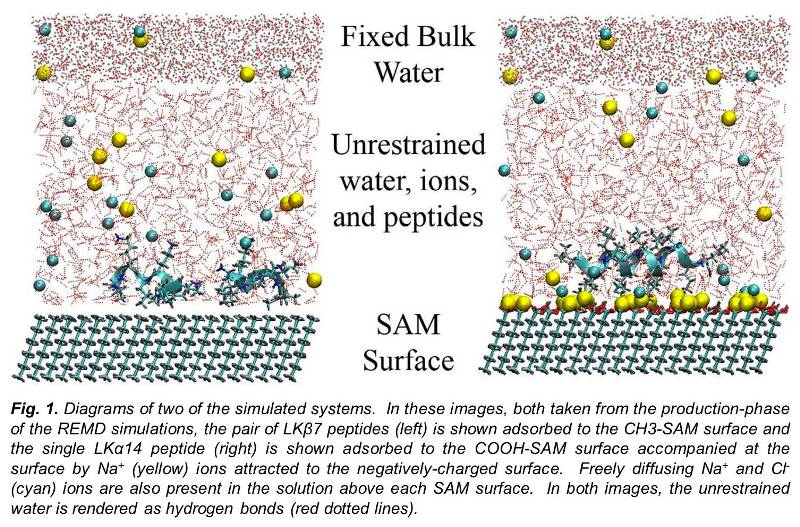 _____________________________________________________________
_____________________________________________________________
NIH/NIGMS
Grant no. R01 GM074511 (sub to Clemson; PI: Castner, Univ. Washington)
