Studies on Biofunctional Polymers Using Advanced Molecular Modeling Methods
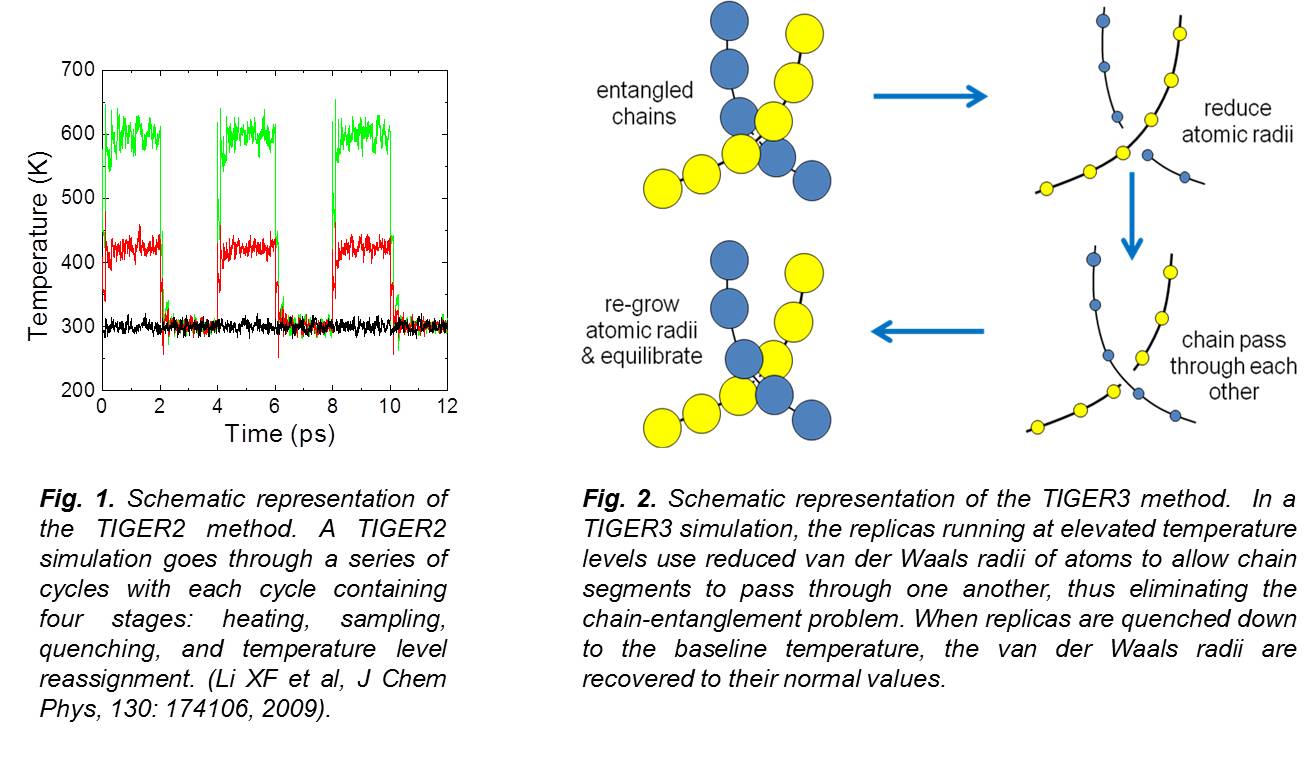
PosterIn this project we were interested in understanding the relationship between structure and function of biofunctional macromolecules. In these studies, we used molecular modeling methods that were proven to be a powerful tool and an excellent complement to experiments in extracting the relationship between structure and function of biofunctional polymers. Computer modeling is able to provide direct observations of the structure and packing at the atomic level and thus provides a direct way to investigate how functional groups influence system behavior. We conducted studies in two directions: (1) developing coarse grained models to simplify complex systems. The coarse graining method provided a means of greatly reducing the number of degrees of freedom in the system while keeping the description of microscopic properties at the necessary level of accuracy and detail; (2) developing advanced modeling methods to accelerate systems sampling and equilibration. We devised the ‘temperature intervals with global exchange of replicas’ (TIGER2) method and the ‘temperature intervals with global exchange of replicas with reduced radii’ (TIGER3) method and validated them by comparison with the replica exchange molecular dynamics (REMD) method for a variety of distinct systems. Compared to the conventional REMD method, our methods required less computational resources while providing higher sampling efficiency.
 _______________________________________________________________
_______________________________________________________________
NIH/NIBIB: RESBIO – Integrated Technologies for Polymer Biomaterials
P41 grant number EB001046 (sub to Clemson, PI: Kohn, Rutgers University)
