Development of Experimental Methods to Study Peptide-Surface Interactions
Poster
The understanding and control of protein adsorption to material surfaces has been one of the major topics of research in the field of biomaterials because of its governing role in cellular response to implants and substrates for biomedical applications. Unfortunately, it has proven to be extremely difficult to quantitatively understand and control these types of interactions. This is because of the complexities involved combined with the fact that methods that previously developed to characterize protein-surface interactions were simply unable to provide the level of detail necessary to achieve this understanding. New, more fundamental methods, both experimental and computational, were needed to overcome these limitations.
At a fundamental level, protein adsorption behavior can be considered to be represented by the combination of the individual interactions between the amino acid residues making up a protein, the solvent environment, and the functional groups presented by a surface. These interactions can be best characterized by the standard-state adsorption free energy (ΔGoads) associated with their adsorption to a functionalized surface, and this information can be potentially very useful in understanding the sub-molecular events that govern protein adsorption behavior.
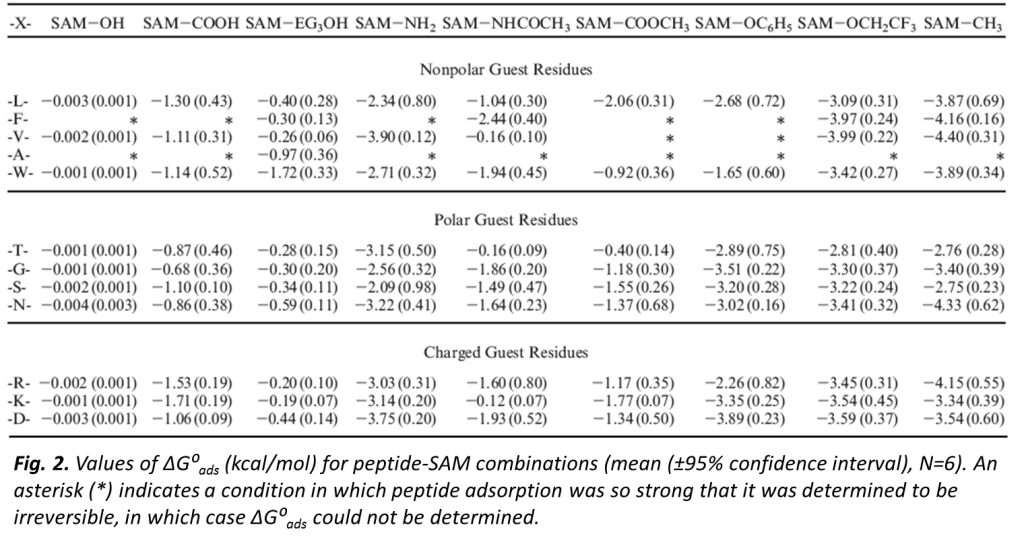
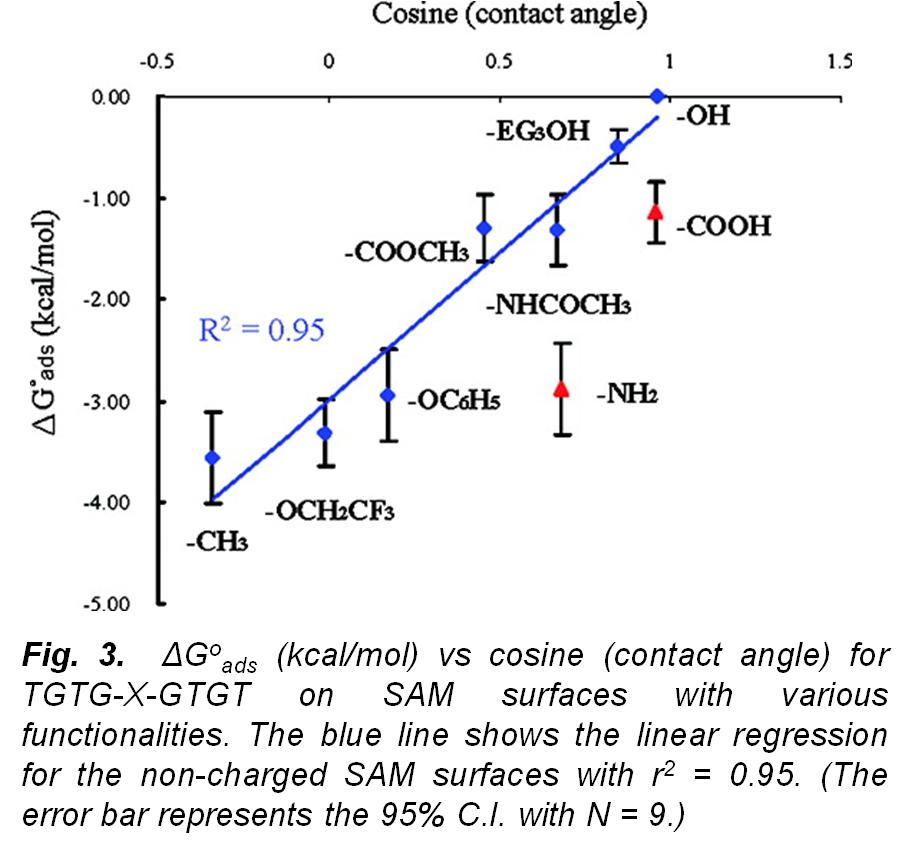
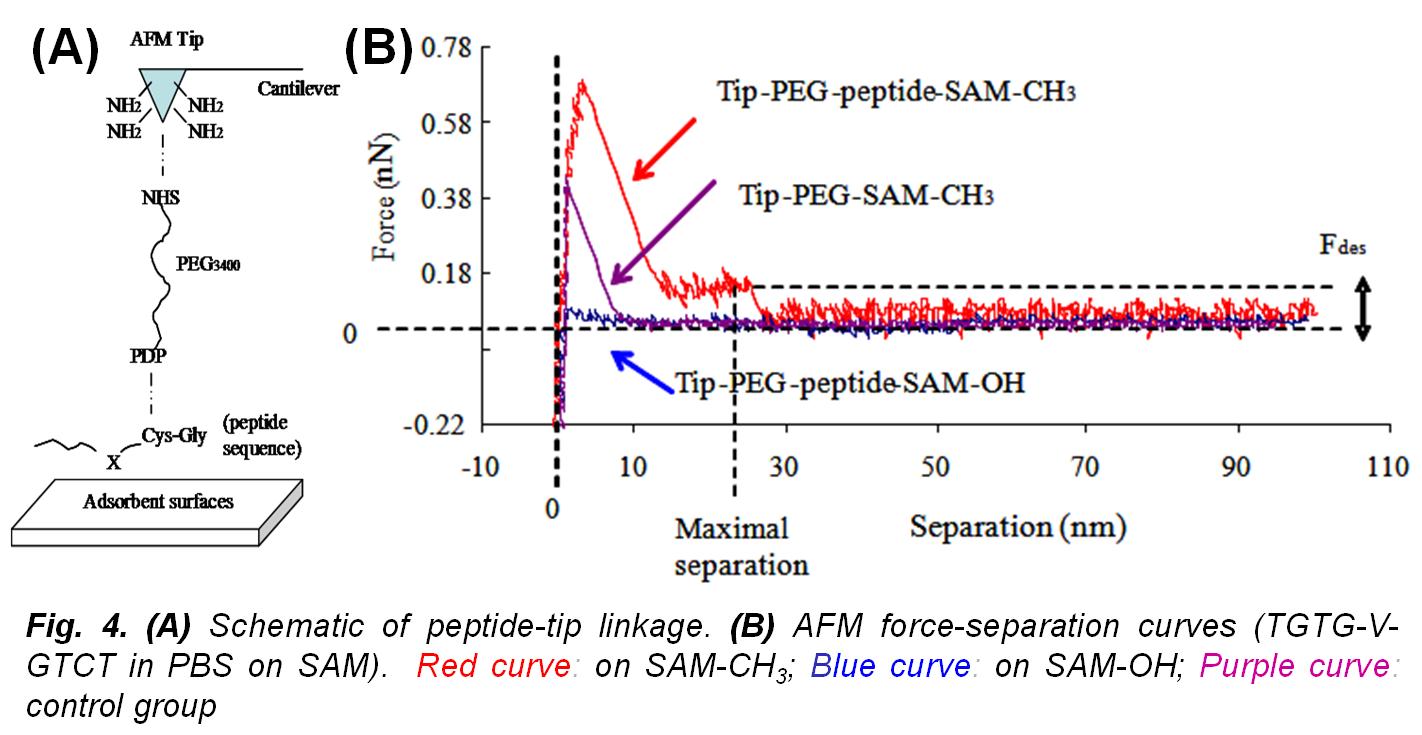
In this research work, 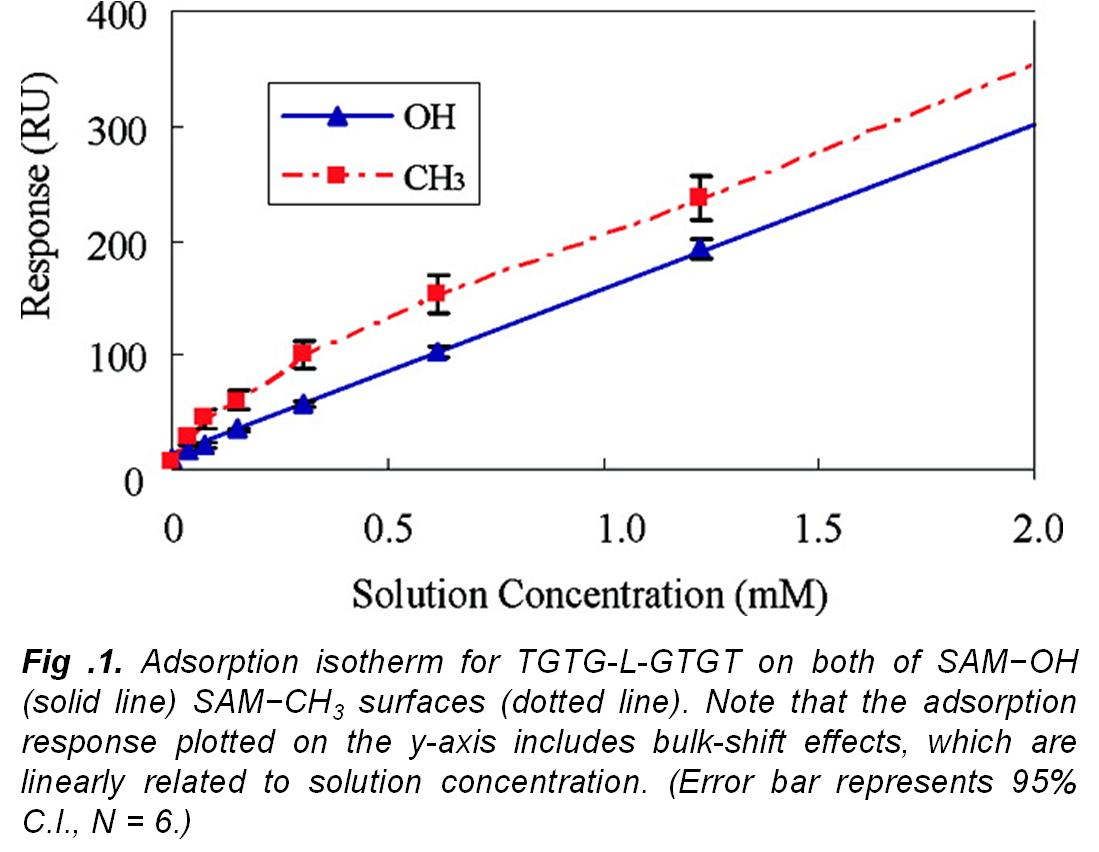 our group developed experimental methods for the determination of (ΔGoads) to quantitatively characterize the peptide adsorption behavior on microscopically flat surfaces presenting functional groups common to many types of polymeric biomaterials using surface plasmon resonance (SPR) spectroscopy and atomic force microscopy (AFM). The development and application of these methods has enabled the fundamental aspects underlying protein adsorption behavior to be characterized and provided data that can be used for the evaluation, modification, and validation of computational models that may be used to accurately predict protein adsorption behavior. As an example of how other groups have used these data, see Bhadra P. and Siu S.W.I., Refined Empirical Force Field to Model Protein-Self-Assembled Monolayer Interactions Based on AMBER14 and GAFF, Langmuir, 35: 9622-9633 (2019).
our group developed experimental methods for the determination of (ΔGoads) to quantitatively characterize the peptide adsorption behavior on microscopically flat surfaces presenting functional groups common to many types of polymeric biomaterials using surface plasmon resonance (SPR) spectroscopy and atomic force microscopy (AFM). The development and application of these methods has enabled the fundamental aspects underlying protein adsorption behavior to be characterized and provided data that can be used for the evaluation, modification, and validation of computational models that may be used to accurately predict protein adsorption behavior. As an example of how other groups have used these data, see Bhadra P. and Siu S.W.I., Refined Empirical Force Field to Model Protein-Self-Assembled Monolayer Interactions Based on AMBER14 and GAFF, Langmuir, 35: 9622-9633 (2019).
______________________________________
Defense Threat Reduction Agency (DTRA)
Grant number HDTRA1-10-1-0028 (PI: Latour)
